Sox2
Uncovering context-dependent transcriptional regulation effects
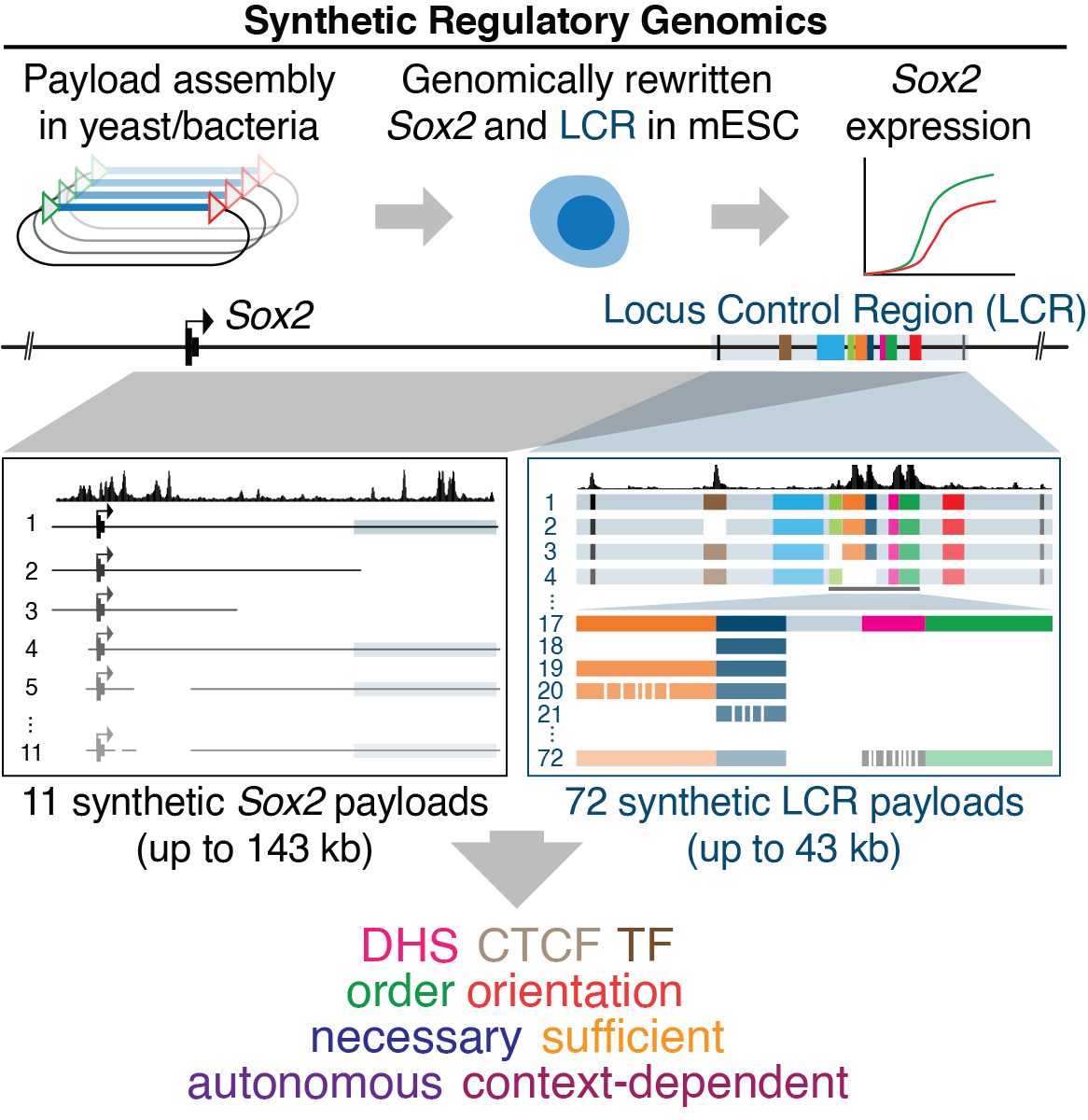
SOX2 is part of a core regulatory network of transcription factors required for embryonic stem cell (ESC) pluripotency maintenance and cellular reprogramming. Expression of Sox2 in mESC depends on a distal cluster of DHSs located ~100 kb away, but their individual contributions and interplay remain elusive. We analyzed the endogenous Sox2 locus using Big-IN to scarlessly integrate large DNA payloads incorporating deletions, rearrangements, and inversions affecting single or multiple DHSs, as well as surgical alterations to transcription factor (TF) recognition sequences. We found that two DHSs comprising a handful of key TF recognition sequences were each sufficient for long-range activation of Sox2 expression. In contrast, three nearby DHSs were entirely context-dependent, showing no activity alone but dramatically augmenting activity of the autonomous DHSs. Our results highlight the role of context as a modulator of genomic regulatory element function, and our synthetic regulatory genomics approach provides a roadmap for dissection of other genomic loci.
Read more in Brosh et al.. Mol Cell 2023
WORKed ON THIS PROJECT:
Ran Brosh, PhD, Institute for Systems Genetics at NYU Langone Health
Matt Maurano, PhD, Institute for Systems Genetics at NYU Langone Health
AND
Andre Mauricio Ribiero Dos Santo, Gwen Ellis, Raquel Ordoñez, Hannah Ashe, Camila Coelho, Megan Hogan, PhD, Angela Hitchcock, Raven Luther, Nicolette Somogyi